Generating 2D diagrams of molecules¶
Introduction¶
The CSD Python API includes functionality for generating 2D diagrams of molecules. It is particularly effective at generating useful 2D diagrams of large complicated molecules including metal-organic compounds.
The diagram generator makes use of a molecular optimisation framework to generate the 2D diagrams. This includes a global optimisation step, which has a stochastic element to it. One is therefore not guaranteed to get the same layout from different runs.
Let us import the ccdc.diagram.DiagramGenerator from the
ccdc.diagram module. Let us also import a
ccdc.io.MoleculeReader to allow us to read in molecules from
the CSD.
>>> from ccdc.diagram import DiagramGenerator
>>> from ccdc.io import EntryReader
Generating 2D diagrams¶
To generate a 2D diagram for a molecule we need to create an instance of
a ccdc.diagram.DiagramGenerator.
>>> diagram_generator = DiagramGenerator()
Let us set the font size to 12 and increase the line width and image size.
>>> diagram_generator.settings.font_size = 12
>>> diagram_generator.settings.line_width = 1.6
>>> diagram_generator.settings.image_width = 500
>>> diagram_generator.settings.image_height = 500
Now let us now read in a moderately complicated compound.
>>> csd_reader = EntryReader('CSD')
>>> mol = csd_reader.molecule('EGEQUD')
To generate a PIL (Python Imaging Library) image of this molecule we call the ccdc.diagram.DiagramGenerator.image() function.
>>> img = diagram_generator.image(mol) # img is a PIL (Python Imaging Library) image
To save this diagram we can use the PIL image’s save() function.
Below are diagrams generated for the CSD entries EGEQUD a ruthenium
porphyrin compound for photodynamic therapy of cancer.

EGEQUD 2D diagram; a ruthenium porphyrin compound for photodynamic therapy of cancer.¶
Accessing diagrams from CSD entries¶
It is possible to get to the 2D diagram stored in the CSD by giving the
ccdc.diagram.DiagramGenerator.image() an ccdc.entry.Entry.
>>> abebuf = csd_reader.entry('ABEBUF')
>>> img = diagram_generator.image(abebuf) # img is a PIL (Python Imaging Library) image
To save this diagram we can use the PIL image’s save() function.
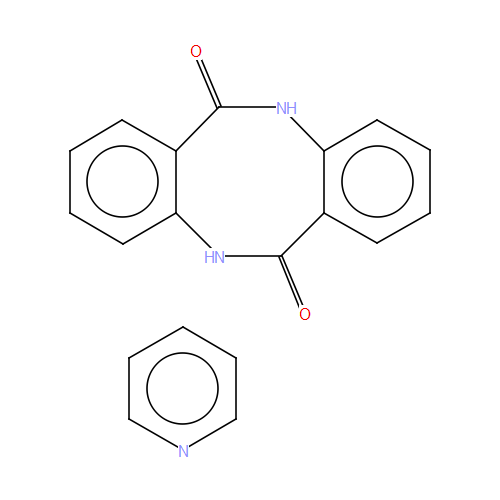
ABEBUF entry 2D diagram.¶
Highlighting atom selections in diagrams¶
It is possible to highlight atoms in a diagram. Let us identify the pyridine using a substructure search. For greater contrast with the rest of the molecule, one can set the default element colouring off:
>>> from ccdc.search import SubstructureSearch, SMARTSSubstructure
>>> searcher = SubstructureSearch()
>>> sub_id = searcher.add_substructure( SMARTSSubstructure('c1ncccc1') )
>>> hits = searcher.search(abebuf.molecule)
>>> selection = hits[0].match_atoms()
>>> diagram_generator.settings.element_coloring = False
>>> img = diagram_generator.image(abebuf, highlight_atoms=selection)
>>> diagram_generator.settings.element_coloring = True # Reset for later

ABEBUF entry 2D diagram with the pyridine ring highlighted.¶
Warning
When highlighting selections in ccdc.molecule.Molecule and
ccdc.crystal.Crystal it is recommended to set the
ccdc.diagram.DiagramGenerator.Settings.shrink_symbols to False
otherwise it may result in misleading diagrams.
For greater contrast with the rest of the molecule, one can set the default element colouring off:
Intra-molecular hydrogen bonds may be displayed. This will require
that ccdc.diagram.DiagramGenerator.Settings.shrink_symbols be set to False. For example:
>>> abahui = csd_reader.molecule('ABAHUI')
>>> diagram_generator.settings.detect_intra_hbonds = True
>>> diagram_generator.settings.shrink_symbols = False
>>> diagram_generator.settings.return_type = 'SVG'
>>> image = diagram_generator.image(abahui)
Note that in this case we have set the return type to ‘SVG’, rather than the default ‘PIL’. This is for illustration only; hydrogen bond depiction will work in either format.

ABAHUI with internal H-bond¶
Diagram generation settings¶
The ccdc.diagram.DiagramGenerator has several settings, which can
be modified. These are stored in a
ccdc.diagram.DiagramGenerator.Settings class.
The default settings are listed below.
>>> diagram_settings = DiagramGenerator.Settings()
>>> print(diagram_settings.line_width)
1.0
>>> print(diagram_settings.image_width)
350
>>> print(diagram_settings.image_height)
350
>>> print(diagram_settings.font_family)
Helvetica
>>> print(diagram_settings.font_weight)
bold
>>> print(diagram_settings.font_italic)
False
>>> print(diagram_settings.font_size)
10
>>> print(diagram_settings.shrink_symbols)
True
>>> print(diagram_settings.explicit_polar_hydrogens)
False
>>> print(diagram_settings.detect_intra_hbonds)
False
>>> print(diagram_settings.overwrite_existing_image)
False
>>> print(diagram_settings.highlight_color)
#ff0000
>>> print(diagram_settings.return_type)
PIL
Substructure Diagrams¶
The diagram generator can create images of instances of ccdc.search.QuerySubstructure or if its derived classes. Highlighting is supported, but the highlight_atoms parameter of the ccdc.diagram.DiagramGenerator.image() method is now an iterable of ccdc.search.QueryAtom instances, or an iterable of ints.
>>> from ccdc import search
>>> smarts_query = search.SMARTSSubstructure('c-c1:c:c:c(-S(=O)(=O)-O):c:c:1')
>>> img = diagram_generator.image(smarts_query)
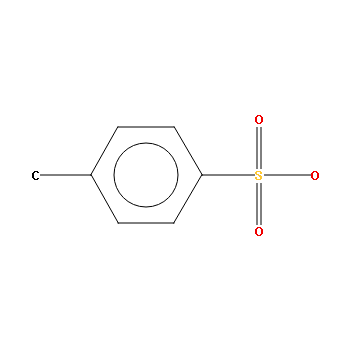
tosyl substructure¶
Generating ChemDraw XML file¶
To generate a ChemDraw XML for a molecule we need to create an instance of
a ccdc.diagram.DiagramGenerator.
>>> diagram_generator = DiagramGenerator()
Let us get a molecule from a SMILES string.
>>> from ccdc.molecule import Molecule
>>> coformer = 'C1CCCCC1'
>>> mol = Molecule.from_string(coformer)
Now let us now get the file to save the ChemDraw XML. Here as an example we create a file in temp dir, then call DiagramGenerator.chemdraw_xml() to write to the file
>>> import tempfile
>>> import pathlib
>>> with tempfile.TemporaryDirectory() as tmpdirname:
... p = pathlib.Path(tmpdirname).joinpath("diag.cdxml")
... cdxml = diagram_generator.chemdraw_xml(mol, p)